Selected Publications
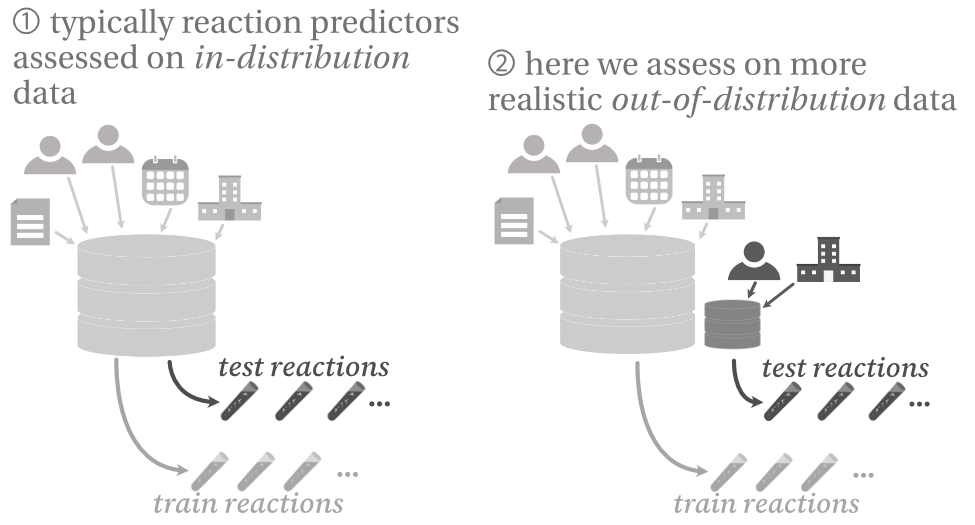
Challenging reaction prediction models to generalize to novel chemistry
preprint, 2025
Despite excellent benchmark performance, ML models for reaction prediction can still struggle on real-world data—we evaluate these limitations by challenging a model on different out-of-distribution tasks.
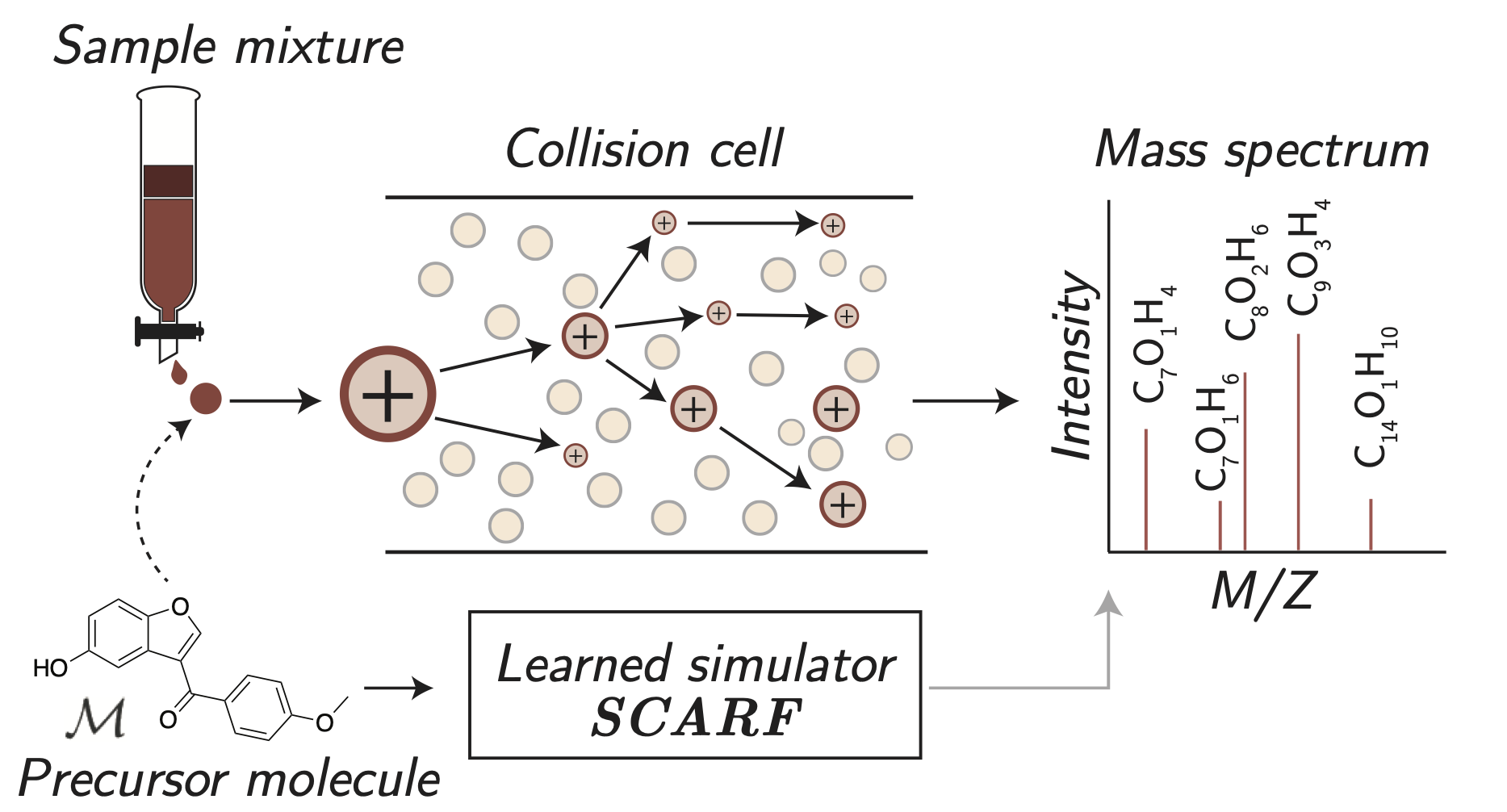
Prefix-tree Decoding for Predicting Mass Spectra from Molecules
Advances in Neural Information Processing Systems 36 (NeurIPS) — Spotlight, 2023
We develop a new method for predicting mass spectra from molecules via first generating the product formulae (the x axis locations of mass spectra peaks) before secondly predicting their intensities (the peaks’ heights on the y axis). In order to overcome the combinatorial explosion in the possible number of product formulae, we design an efficient and compact representation for these formulae based on prefix trees.
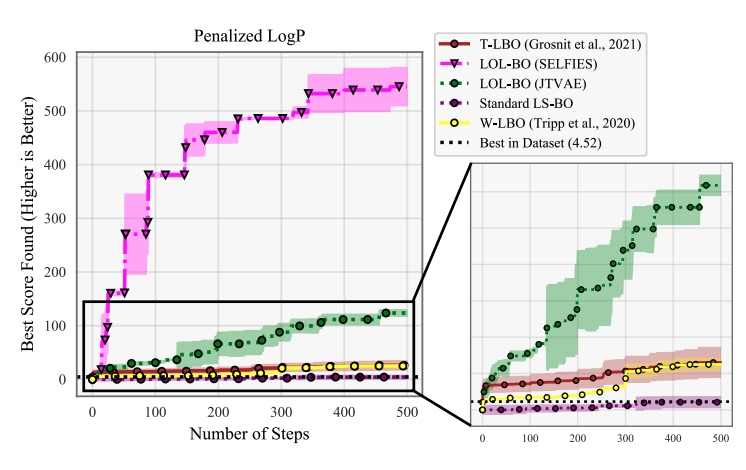
Local Latent Space Bayesian Optimization over Structured Inputs
Advances in Neural Information Processing Systems 35 (NeurIPS), 2022
We develop LOL-BO, an algorithm to more efficiently perform latent space optimization (optimization over hard discrete spaces by transforming them into continuous problems) by recognizing latent space optimization is still high dimensional optimization.

Machine Learning Methods for Modeling Synthesizable Molecules
PhD Thesis, 2021
My PhD thesis, which brings together some of the papers listed below to describe physically inspired machine learning models for reaction prediction and de novo design.
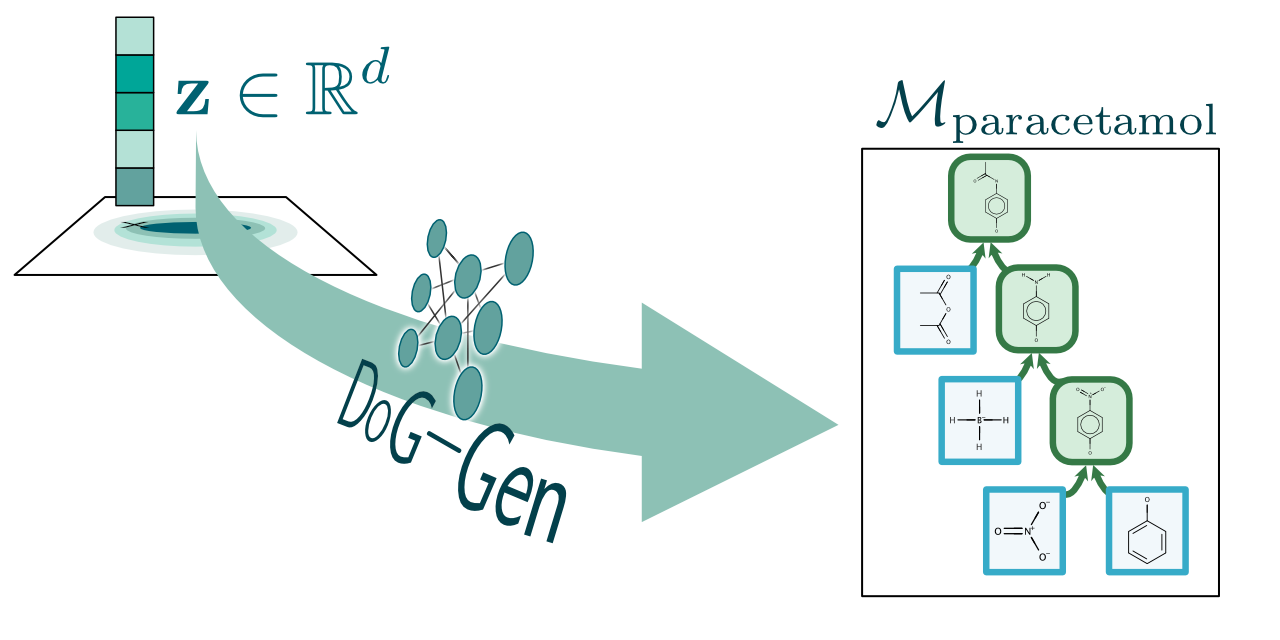
Barking up the right tree: an approach to search over molecule synthesis DAGs
Advances in Neural Information Processing Systems 33 (NeurIPS) — Spotlight, 2020
We describe how complex synthesis plans can be described using molecule synthesis DAGs and design a generative model over this structure. Not only can our model find molecules with good properties, but those that are synthesizable and stable too, allowing the unconstrained optimization of an inherently constrained problem.
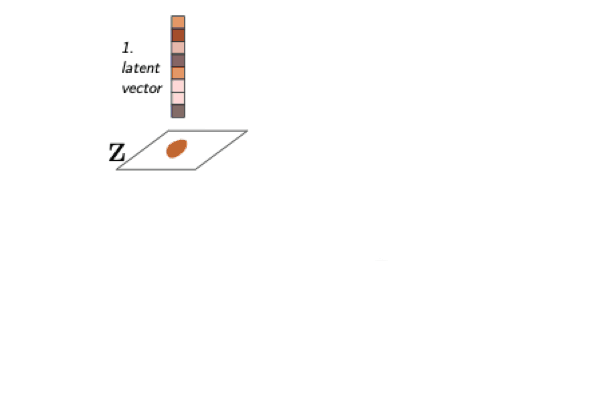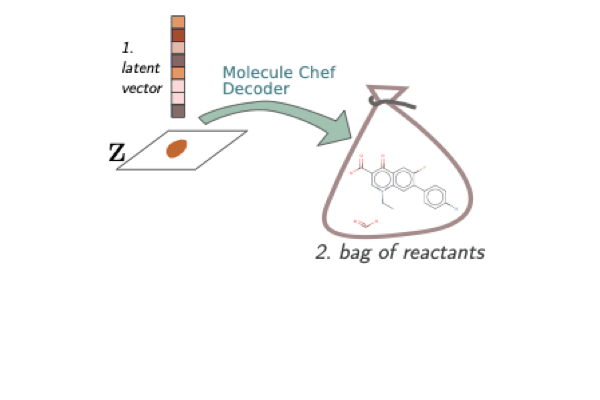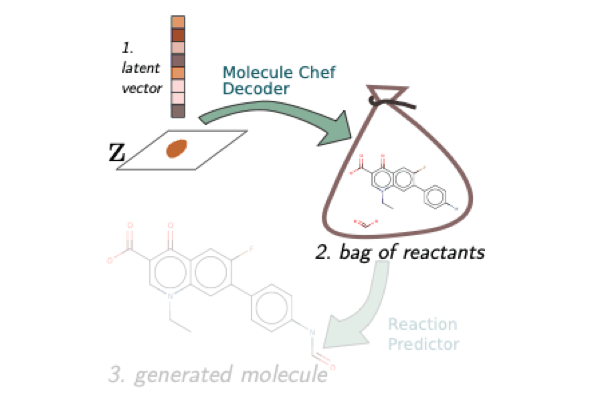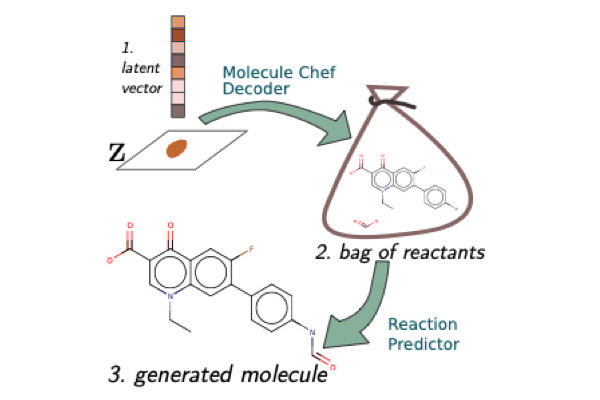
A Model to Search for Synthesizable Molecules
Advances in Neural Information Processing Systems 32 (NeurIPS), 2019
When designing a new molecule, you not only want to know what to make, but, critically, how to make it! We propose a deep generative model for molecules (Molecule Chef) that provides this information, by first constructing a set of reactants before reacting them together to form a final molecule.
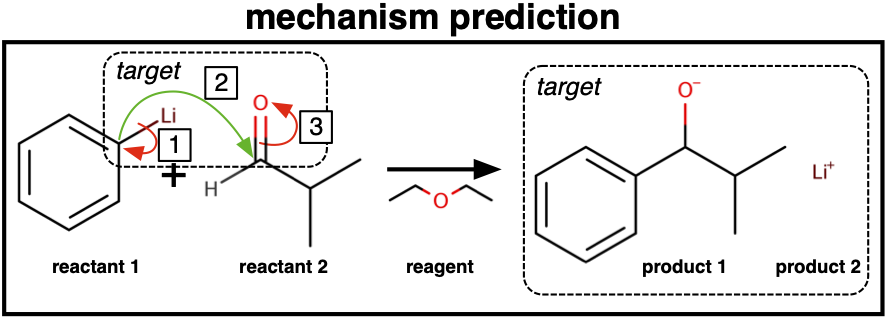
Are Generative Classifiers More Robust to Adversarial Attacks?
International Conference on Machine Learning (ICML), 2019
We propose and analyze the deep Bayes classifier, an extension of naive Bayes using deep conditional generative models. We show that these models (a) are more robust to adversarial attacks than deep discriminative classifiers, and (b) also allow the principled construction of attack detection methods.
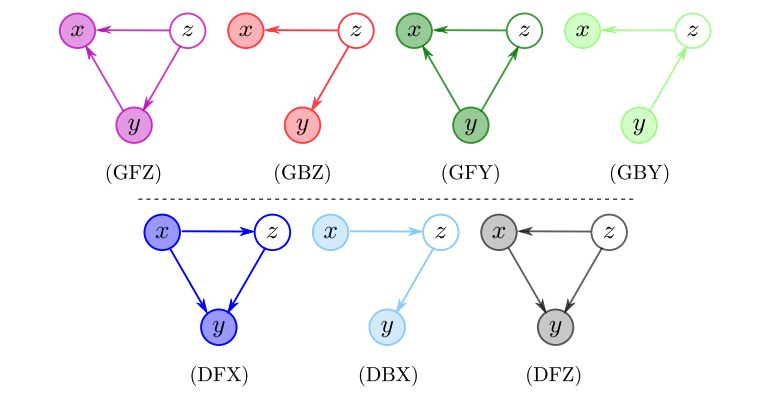
A Generative Model For Electron Paths
International Conference on Learning Representations (ICLR), 2019
We devise a method to model a wide class of common reactions through parametrizing movements of electrons, breaking down the modeling of a reaction into simple, intuitive steps. We come up with an approach to generate approximate electron movements from atom-mapped reaction datasets and show how this can be used to train our model.
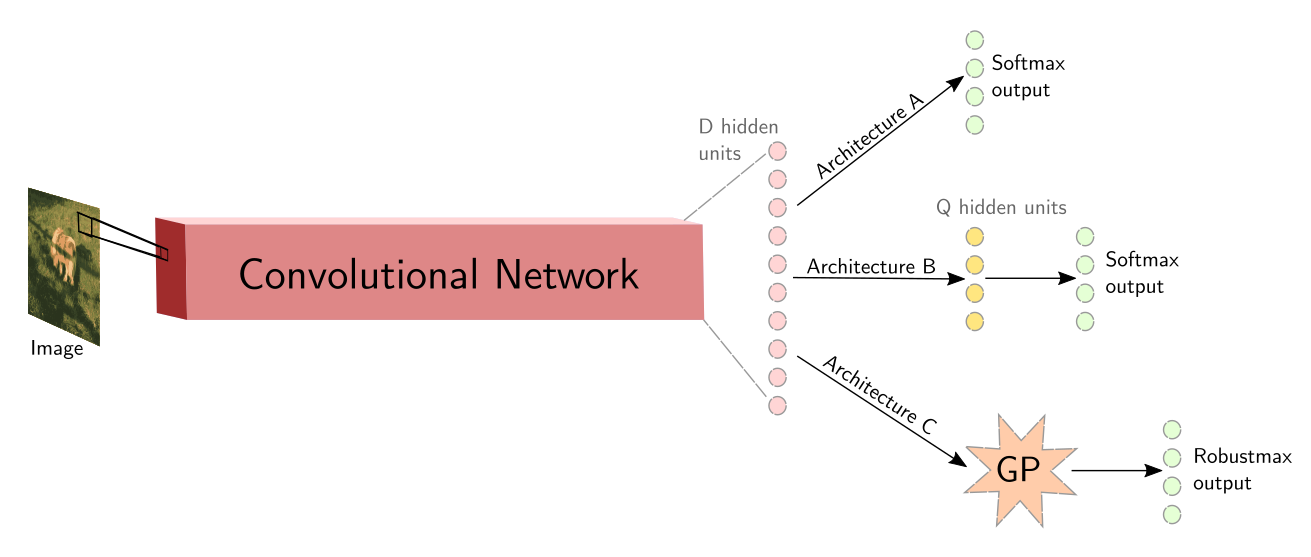
Adversarial Examples, Uncertainty, and Transfer Testing Robustness in Gaussian Process Hybrid Deep Networks
Reliable Machine Learning in the Wild - ICML 2017 Workshop, 2017
We look at the properties of GPDNNs, Gaussian processes composed with deep neural networks. We show that unlike regular feedforward neural networks, GPDNNs often know when they don't know. This means that they often avoid making overconfident predictions on out of domain data.